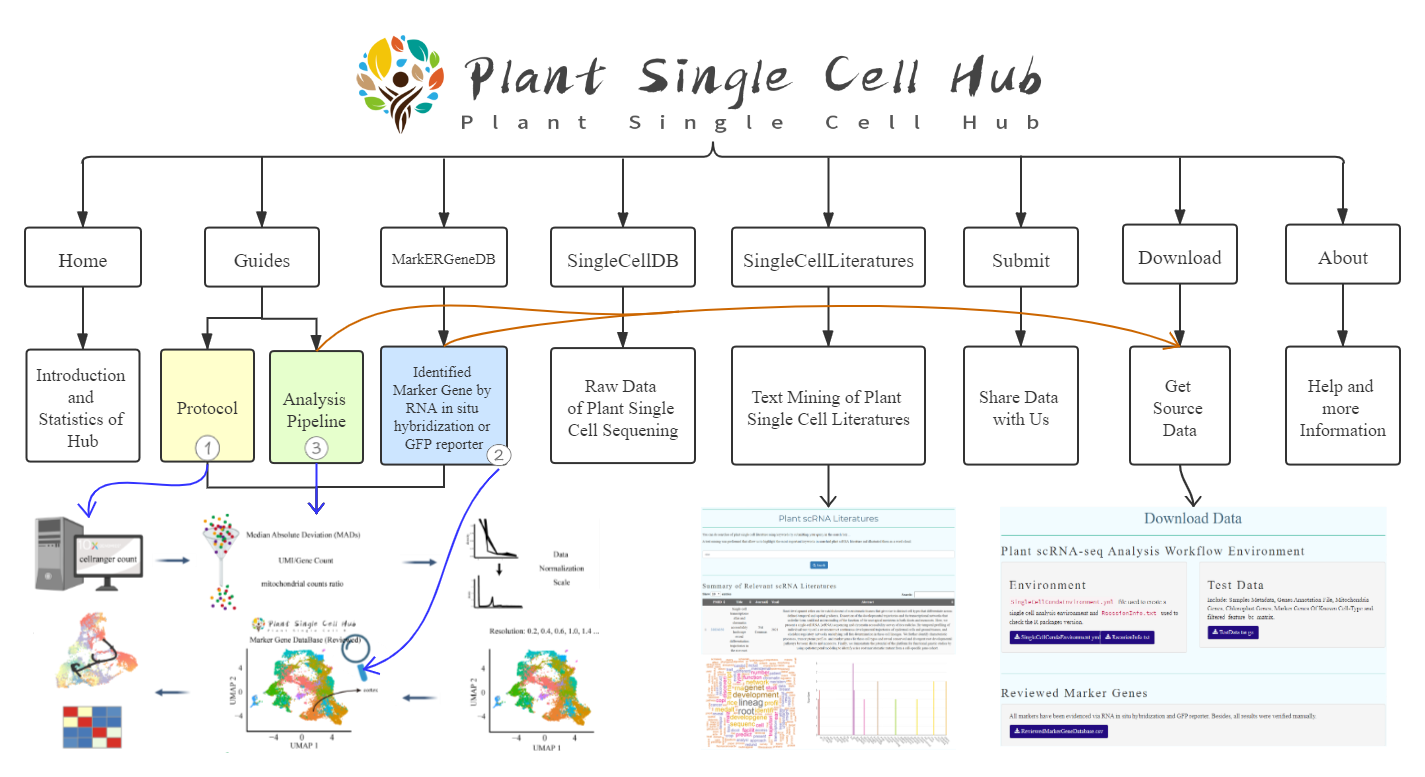
What can we do?
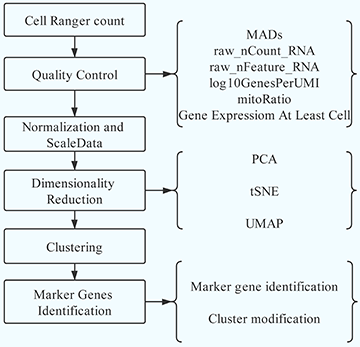
We are provided a workflow to analyze plants Single cell RNA-seq data, which includes quality control based on raw_nCount_RNA, raw_nFeature_RNA, Mitochondria Genes and Chloroplast Genes.
VIEW WORKFLOWWe are provided a plant marker genes database that all genes have been evidenced via RNA in situ hybridization or GFP reporter.
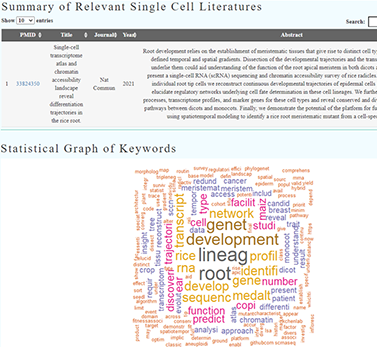
VIEW DATABASEWe are providing an interface that can more convenient to get the literature of plant single cell and uses natural language processing (NLP) to try mine the new information or help answer specific research questions.
VIEW LITERATURESHow to prepare input data for workflow?
The gene id should be have same format in follows files. Especially for Cell Ranger Count Matrix.
1. Cell Ranger Count Data
The filtered_feature_bc_matrix need input into analysis workflow can be count follows cellranger mkref and cellranger count. More detail can be find in Running cellranger count.
2. Samples Metadata
A comma separated csv format file that used to describe the information of samples. It's should been have the header: library_id, molecule_h5, batch, AMLStatus and condition.
A example file can be found here: Sample_MetaData.csv
| library_id | molecule_h5 | batch | AMLStatus | condition |
|---|---|---|---|---|
| T0_1 | Cell_Ranger/T0_1/outs/molecule_info.h5 | v3_lib | T0 | treatment |
| T0_2 | Cell_Ranger/T0_2/outs/molecule_info.h5 | v3_lib | T0 | treatment |
| T1_1 | Cell_Ranger/T1_1/outs/molecule_info.h5 | v3_lib | T1 | control |
| T1_2 | Cell_Ranger/T1_2/outs/molecule_info.h5 | v3_lib | T1 | control |
3. Genes Annotation File
A tab separated txt format file that includes the annotation of gene function. It's should be no header.
A example file can be found here: Genome_Genes_Function_Annotation.txt
| Ghir-A01G000050 | AT4G21192.1 | 3.00E-44 | Symbols: | Cytochrome c oxidase biogenesis protein Cmc1-like | chr4:11294563-11295192 FORWARD LENGTH=80 |
| Ghir-A01G000040 | AT1G64260.1 | 2.00E-39 | Symbols: | MuDR family transposase | chr1:23847756-23849915 FORWARD LENGTH=719 |
4. Mitochondria Genes
A csv format file that includes the mitochondria genes. It's should have the header: gene_id.
A example file can be found here: MitochondriaGenes.csv
| gene_id |
|---|
| Ghir-D13G022910 |
| Ghir-D09G012400 |
5. Chloroplast Genes
A csv format file that includes the chloroplast genes. It's should have the header: gene_id.
A example file can be found here: ChloroplastGenes.csv
| gene_id |
|---|
| Ghir-D08G003430 |
| Ghir-A05G030920 |
6. Marker Genes Of Known Cell-Type
A comma separated csv format file that includes the marker genes and cell annotation. It's should have the header: name, parent and genes.
Multiple marker genes for a cell cluster should be list in different row.
A example file can be found here: Maker_SC.csv
| name | parent | genes |
|---|---|---|
| cortex | cortex | Ghir-A03G023470 |
| cortex | cortex | Ghir-A03G023570 |
| metaphloem sieve element | phloem | Ghir-A05G017430 |
| metaxylem | xylem | Ghir-A02G012540 |
How to run workflow?
We are provides a Single Cell Conda Environment and R Session Infomation: SingleCellCondaEnvironment.yml and RsessionInfo.txt.
You can easily configure an environment used to run this workflow.👇
conda env update --file SingleCellCondaEnvironment.yml --prune
Firstly, you should have a conda environment. If not, please install it.
Why not provide online scRNA analysis services?
We are based on the following two considerations: plants Single cell RNA data is really big data and extreme complexity.
We need to combine species information, single cell sample status and the corresponding biological background to adjust the parameters repeatedly.
Fixed bioinformatics calculation process can not get ideal results.
Therefore, we customize the calculation process and provide download by specifying key parameters. Users can perform multiple analyses on their own servers.
How to submit data or facrback question with us?
Don't be shy.😳 xzp@mail.hzau.edu.cn
Updated History/Release Information
2025-07-03
Updated 2021-12-10
Updated 2021-11-3
Updated 2021-05-23
External Links
scRNA-tools database is a catalogue of tools for analysing single-cell RNA sequencing data
VIEW TOOLSPlantscRNAdb is a database for plant single-cell RNA analysis (Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum and Zea mays).
VIEW DATABASESingle Cell Expression Atlas is unique in that it provides to the life sciences community uniformly analysed and annotated single cell RNA-Seq data across multiple species
VIEW ATLASPlantCellMarker is unique in that it provides to the life sciences community uniformly analysed and annotated single cell RNA-Seq data across multiple species
VIEW PCMDBhuman Ensemble Cell Atlas (hECA) provides a platform for assembling massive scattered single-data into a unified Giant Table (uGT).
VIEW TOOLS