1. Introduction
Welcome to T2TCotton-Hub, an integrative database for functional genomics data of high efficient regenerated cotton Jin668 and YZ1. In T2TCotton-Hub you can:
- Explore and search genomic features, including coding sequences and transposons.
- Insight into collinearity between genomes.
- CRISPR sgRNA design and high throughput detection of editing plants.
- Explore the changes of gene expression in the early stage of somatic embryogenesis at the single cell level.
- Explore gene expression and regulatory networks among different tissues.
- Bioinformatics analysis of genes or gene sets of interest to you.
- Download pre-calculated genomics data for further analysis.
2. Cotton Species and Regeneration
The Gossypium genus contains about 50 speciesWendel J F, Albert V A. Phylogenetics of the cotton genus (Gossypium): character-state weighted parsimony analysis of chloroplast-DNA restriction site data and its systematic and biogeographic implications [J]. Syst Bot, 1992: 115-143.. Most of them are diploids and can be grouped to 8 diploid members (marked as A~G and K). They have different geographical origins and variable estimated genome sizes, mostly due to different amount of DNA repeat elements.
In cotton genetic transformation, only very limited genotypes, such as Coker 201, 310, 312, SM3, ZM24Yang, Z. et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nature Communications 10, 2989 (2019)., YZ1Li, J. et al. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnology Journal 17, 435-450 (2019). and Jin668Li, J. et al. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnology Journal 17, 435-450 (2019). could achieve plant regeneration through somatic embryogenesis. Among them, Jin668 was a newly developed elite genotype with extremely high plant regeneration ability by our team through Successive Regeneration Acclimation (SRA) strategyLi, J. et al. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation process. Plant Biotechnology Journal 17, 435-450 (2019)., which has been widely applied in more than fifty cotton research institutes around the world and recognized by International Cotton Genome Initiative (ICGI, 2021 Cotton Biotechnology Award receptor-Dr. Shuangxia Jin).

2.1 TM-1
G. hirsutum L. acc. Texas Marker-1 (TM-1), which is widely used as a genetic standard with no-regeneration abilityChen, Z.J. et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145, 1303–1310 (2007)..
2.2 YZ1
G. hirsutum L. acc. YZ1, a Upland cotton genotype with high regeneration ability.
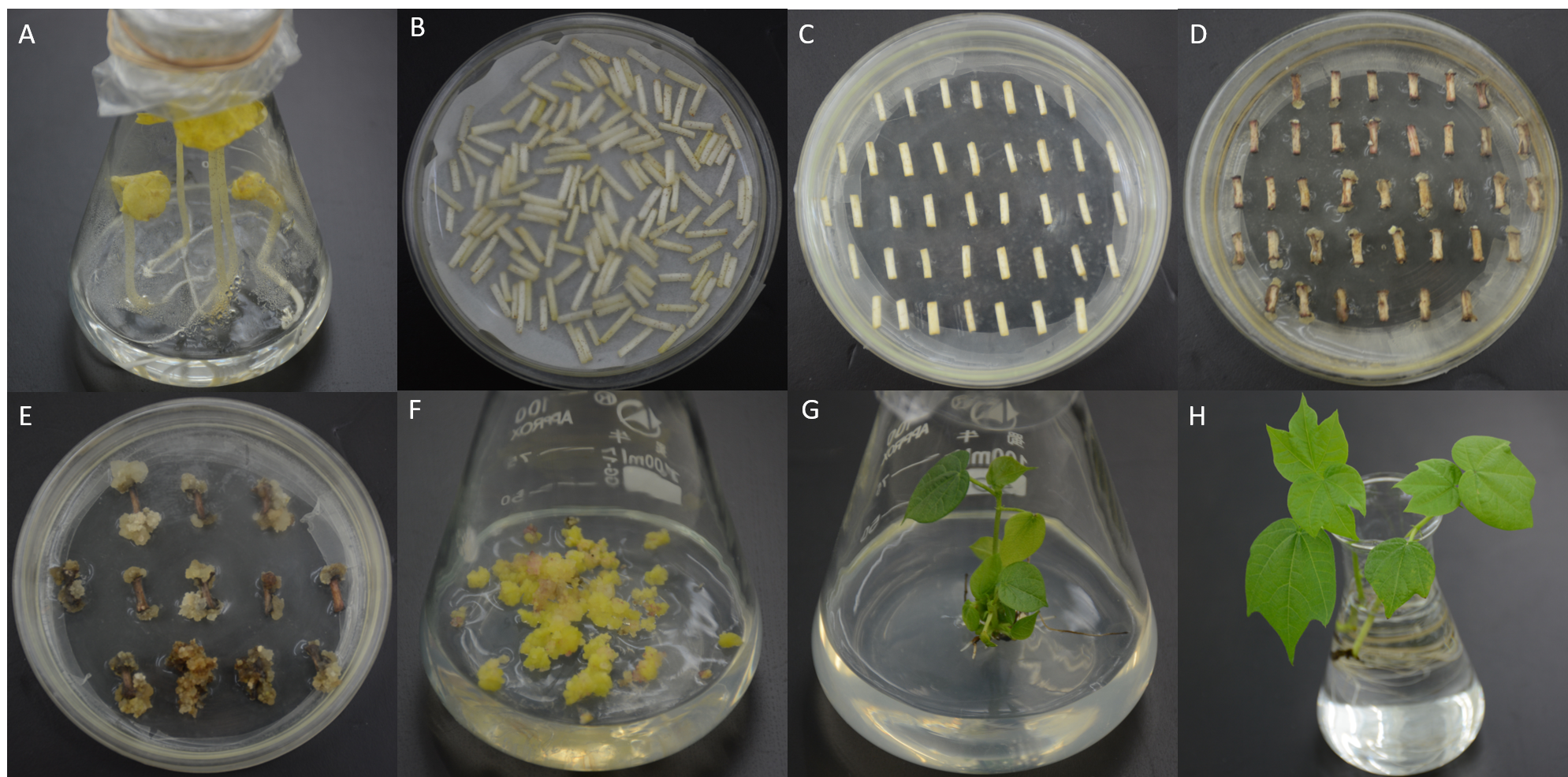
2.3 Jin668
G. hirsutum L. acc. Jin668, a newly developed elite genotype with extremely high plant regeneration ability by our team through Successive Regeneration Acclimation (SRA) strategy.

3. Genome
Genome Assembly
| genotype | Information |
|---|---|
Jin668 |
T2T-Jin668; http://jinlab.hzau.edu.cn/T2TCottonHub/download/ |
YZ1 |
YZ1; http://jinlab.hzau.edu.cn/T2TCottonHub/download/ |
TM-1 |
Ghirsutum_genome_HAU_v1.1.tar.gz; http://cotton.hzau.edu.cn/EN/Download.htm |
Gene Models
| genotype | Gene ID | Transcript ID |
|---|---|---|
Jin668 |
Ghjin_D01g000020 | Ghjin_D01g000020.1 |
YZ1 |
Ghyz_D01g000020 | Ghyz_D01g000020.1 |
TM-1-v3.1 |
Gohir.A01G001500 | Gohir.A01G001500 |
TM-1-HZAU |
Ghir_A01G000010 | Ghir_A01G000010.1 |
4. CRISPR Hub
Barcode Primers Design
This program generates barcodes of a desired length, distance, and GC content. First, your need the base primer paired end primers.
| Primers (5'to3') | Seq |
|---|---|
1_PGF-crRNA2-F_aaaatc |
aaaatcTTGGCATAGTGCTGATGGAAG |
1_PGF-crRNA2-R_aaaatc |
gattttAGCCACGATTCCAAGGATTAA |
5. Single Cell Hub
Cell Landscape in Cotton (CLC)
A single cell landscape in cotton.
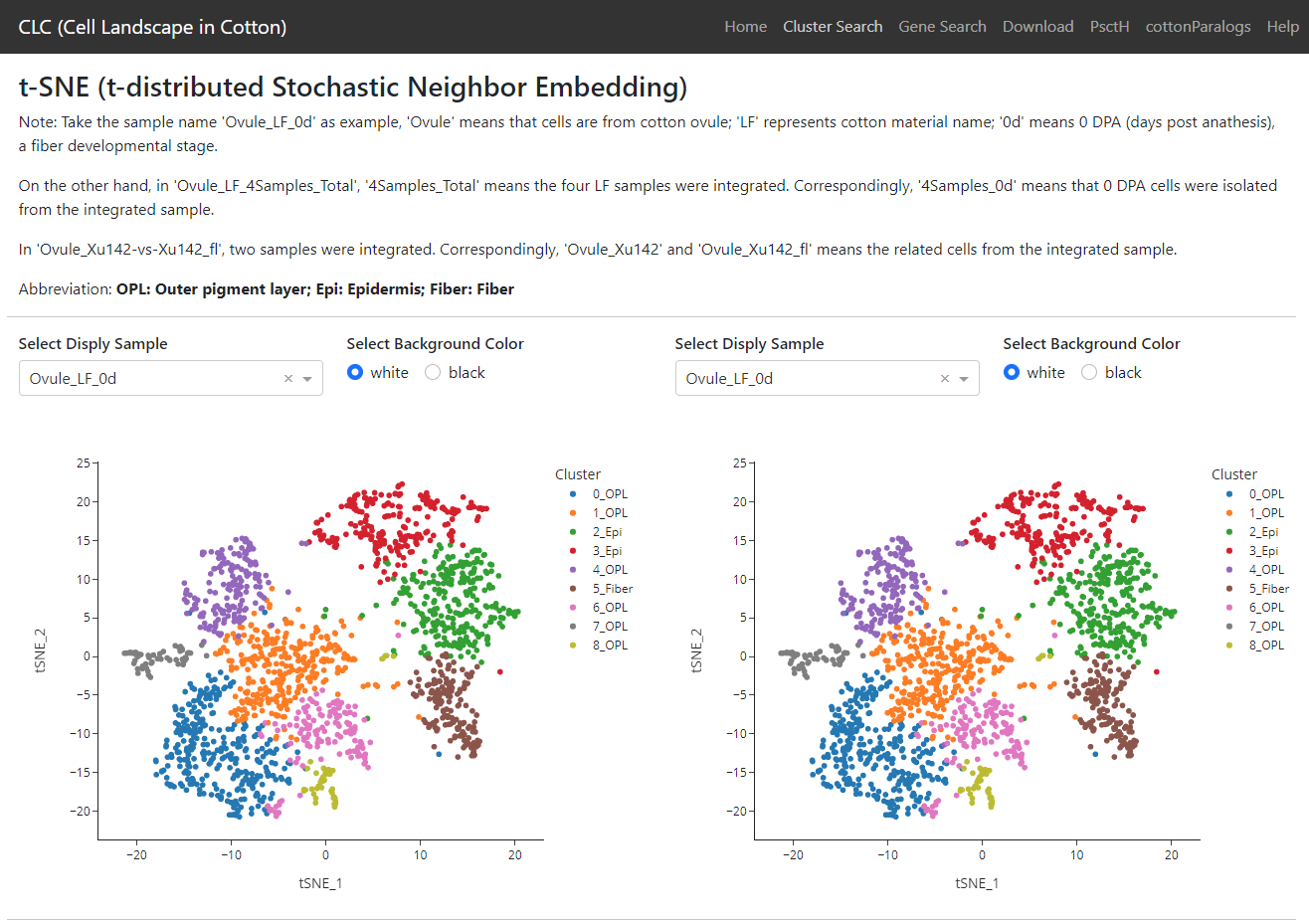
Plant Single Cell Hub (PsctH)
An integrated online tool to explore the plant single-cell transcriptome landscape.

6. Expression
As shown in the right figure, we performed RNA-seq on different tissues of Jin668. The RNA sequencing reads were removed of adapters and trimmed for low-quality bases using Trimmomatic (v0.39) (Bolger et al., 2014). The clean reads were then mapped to the reference genome using HISAT2 (v2.2.1) (Kim et al., 2015) with default parameters. The expression level (transcripts per million; TPM) of genes was calculated by StringTie (v2.1.4) (Pertea et al., 2015).
Multiple Gene Expression
Input one or more gene IDs and then select the tissue type to obtain the expression level and heatmap of the selected genes.
House-keeping & Tissue-specific Genes
By calculated the general scoring metric Tau specificity index (Lüleci and Yılmaz, 2022), the tissue-specific expression genes were obtained in Jin668.
7. Tools
Gene Information
In the model, we collected > 75,854 genes information of cotton. Users can got them by searching gene locus, gene symbol or related function. The result will be showed in a table and we recommend click each link to know more knowledge detail for interested genes.
Gene ID Conversion
Use Reciprocal best hits BLAST (RBHB) to convert genomic coordinates between TM-1, Jin668 and YZ1 for a group of genes.
Phylogenetic Tree
Use muscle and FastTree to alignment and build the phylogenetic tree for a group of genes.
Chromosome Mapping
Use karyoploteR to visualize the relative position of a set of genes on chromosomes.
Blast
Quickly obtain the homeotic gene of other species compared with Jin668.
Sequence Fetch
Sequence Fetch is a model of Retrieving sequences in T2TCotton-Hub. To export gene sequence, click the 'Fetch' button after confirmed coordinate or locus id. At the same time, you will get the mRNA and protein sequences.
Bulk Data Retrieval
This page allows you to retrieve genomic information in bulk, including protein or nucleotide sequences of candidate genes.
Literature Mining
Quickly search for research literature in the field of cotton.
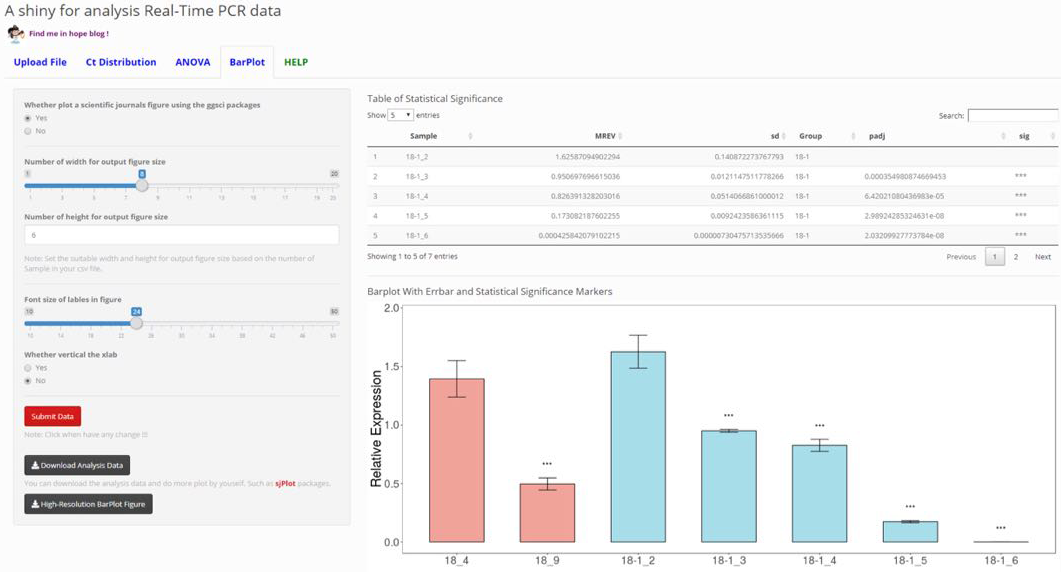
qRT-PCR Analysis
Auto-analysis for Real-Time PCR (qPCR) data.
8. Download
Provide genome assembly statistics and corresponding genome files for T2T-Jin668 and YZ1.