Our Publications
Background
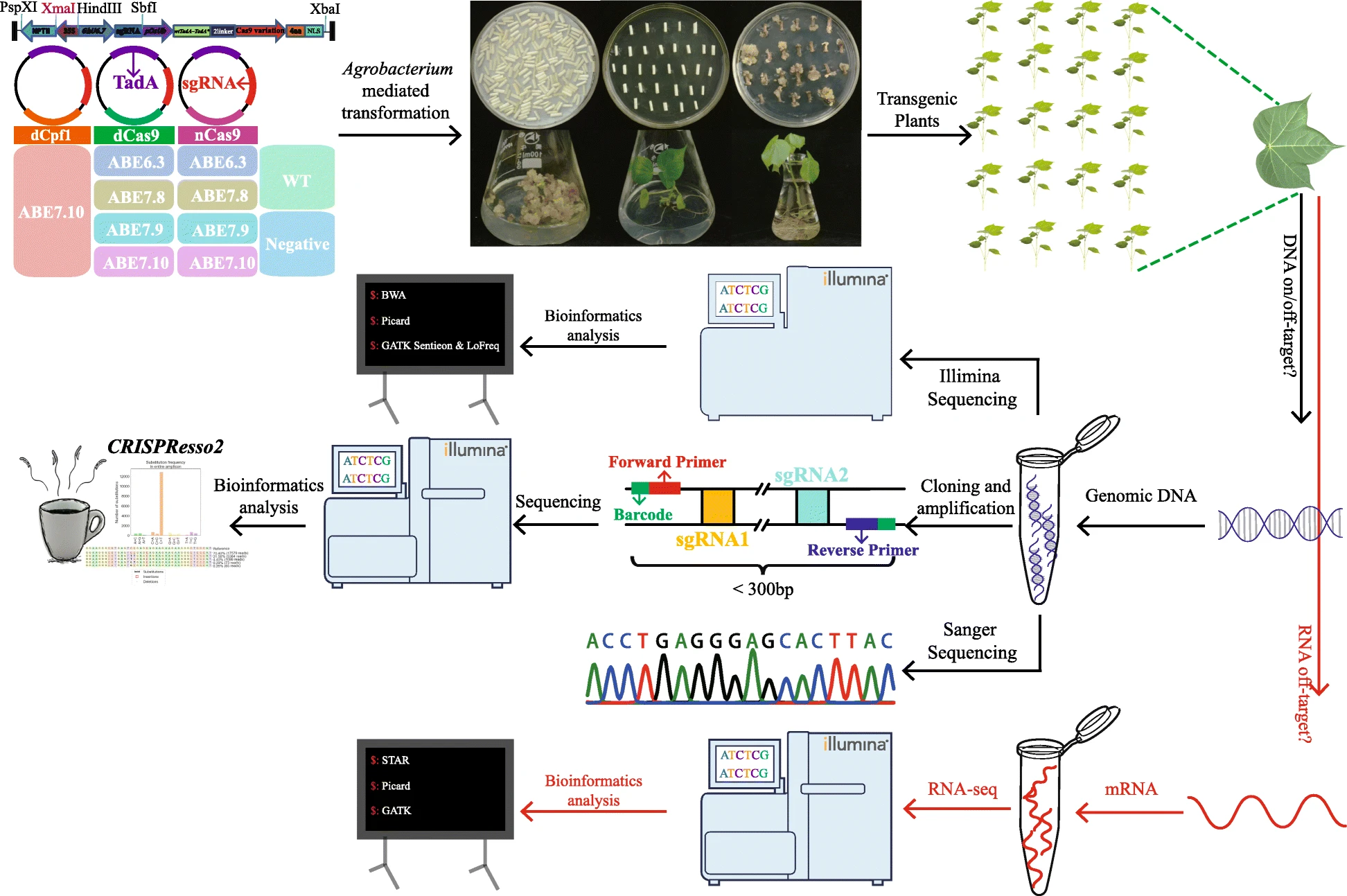
Base editors (BEs) display diverse applications in a variety of plant species such as Arabidopsis, rice, wheat, maize, soybean, and cotton, where they have been used to mediate precise base pair conversions without the collateral generation of undesirable double-stranded breaks (DSB). Studies of single-nucleotide polymorphisms (SNPs) underpinning plant traits are still challenging, particularly in polyploidy species where such SNPs are present in multiple copies, and simultaneous modification of all alleles would be required for functional analysis. Allotetraploid cotton has a number of homoeologous gene pairs located in the A and D sub-genomes with considerable SNPs, and it is desirable to develop adenine base editors (ABEs) for efficient and precise A-to-G single-base editing without DSB in such complex genome.
Results
We established various ABE vectors based on different engineered adenosine deaminase (TadA) proteins fused to Cas9 variants (dCas9, nCas9), enabling efficient A to G editing up to 64% efficiency on-target sites of the allotetraploid cotton genome. Comprehensive analysis showed that GhABE7.10n exhibited the highest editing efficiency, with the main editing sites specifically located at the position A5 (counting the PAM as positions 21–23). Furthermore, DNA and RNA off-target analysis of cotton plants edited with GhABE7.10n and GhABE7.10d by whole genome and whole-transcriptome sequencing revealed no DNA off-target mutations, while very low-level RNA off-target mutations were detected. A new base editor, namely GhABE7.10dCpf1 (7.10TadA + dCpf1), that recognizes a T-rich PAM, was developed for the first time. Targeted A-to-G substitutions generated a single amino acid change in the cotton phosphatidyl ethanolamine-binding protein (GhPEBP), leading to a compact cotton plant architecture, an ideotype for mechanized harvesting of modern cotton production.
Conclusions
Our data illustrate the robustness of adenine base editing in plant species with complex genomes, which provides efficient and precise toolkit for cotton functional genomics and precise molecular breeding.
Summary
The base‐editing technique using CRISPR/nCas9 (Cas9 nickase) or dCas9 (deactivated Cas9) fused with cytidine deaminase is a powerful tool to create point mutations. In this study, a novel G. hirsutum‐Base Editor 3 (GhBE3) base‐editing system has been developed to create single‐base mutations in the allotetraploid genome of cotton (Gossypium hirsutum). A cytidine deaminase sequence (APOBEC) fused with nCas9 and uracil glycosylase inhibitor (UGI) was inserted into our CRISPR/Cas9 plasmid (pRGEB32‐GhU6.7). Three target sites were chosen for two target genes, GhCLA and GhPEBP, to test the efficiency and accuracy of GhBE3. The editing efficiency ranged from 26.67 to 57.78% at the three target sites. Targeted deep sequencing revealed that the C→T substitution efficiency within an ‘editing window’, approximately six‐nucleotide windows of −17 to −12 bp from the PAM sequence, was up to 18.63% of the total sequences. The 27 most likely off‐target sites predicted by CRISPR‐P and Cas‐OFFinder tools were analysed by targeted deep sequencing, and it was found that rare C→T substitutions (average < 0.1%) were detected in the editing windows of these sites. Furthermore, whole‐genome sequencing analyses on two GhCLA‐edited and one wild‐type plants with about 100× depth showed that no bona fide off‐target mutations were detectable from 1500 predicted potential off‐target sites across the genome. In addition, the edited bases were inherited to T1 progeny. These results demonstrate that GhBE3 has high specificity and accuracy for the generation of targeted point mutations in allotetraploid cotton.

Summary
The results presented demonstrate that 34 °C is the optimum temperature for active CRISPR/LbCpf1 in cotton, and the bleached, glandless phenotype facilitates analysis. More importantly, homozygous, nontransgenic and gossypol‐free plants provide valuable new germplasm for molecular breeding programs. The opportunity is demonstrated for improved editing efficiency in cotton by simple heat treatment during seed sowing or plant tissue culture.

Summary
The CRISPR/Cas9 and Cas12a (Cpf1) tools have been used on a large scale for genome editing. A new effector with a single nuclease domain, a relatively small size, low‐frequency off‐target effects and cleavage capability under high temperature has been recently established and designated CRISPR/Cas12b (C2c1). Cas12b has also shown temperature inducibility in mammalian systems. Therefore, this system is potentially valuable for editing the genomes of plant species, such as cotton, that are resistant to high temperatures. Using this new system, mutants of upland cotton were successfully generated following Agrobacterium‐mediated genetic transformation under a range of temperatures. Transformants (explants infected by Agrobacterium) exposed to 45 °C for 4 days showed the highest editing efficiency. No off‐target mutation was detected by whole‐genome sequencing. Genome edits by AacCas12b in T0 generation were faithfully passed to the T1 generation. Taken together, CRISPR/Cas12b is therefore an efficient and precise tool for genome editing in cotton plants.
